Mito Clones¶
Using vireo for clonal reconstruction - mitochondrial mutations
Date: 15/03/2022
The mitochondrial mutations data set is extracted from Ludwig et al, Cell, 2019, the 9 variants used here are from Supp Fig. 2F (and main Fig. 2F).
Generally, you can use cellSNP-lite to genotype mitochondrial genomes and call clonal informed mtDNA variants with MQuad.
With the filtered variants at hand, we can use the vireoSNP.BinomMixtureVB class to reconstruct the clonality.
[1]:
import vireoSNP
import numpy as np
from scipy import sparse
from scipy.io import mmread
import matplotlib.pyplot as plt
print(vireoSNP.__version__)
0.5.6
[2]:
# np.set_printoptions(formatter={'float': lambda x: format(x, '.5f')})
[3]:
AD = mmread("../data/mitoDNA/cellSNP.tag.AD.mtx").tocsc()
DP = mmread("../data/mitoDNA/cellSNP.tag.DP.mtx").tocsc()
# mtSNP_ids = np.genfromtxt('../data/mitoDNA/passed_variant_names.txt', dtype='str')
Identify clones¶
[4]:
from vireoSNP import BinomMixtureVB
[5]:
_model = BinomMixtureVB(n_var=AD.shape[0], n_cell=AD.shape[1], n_donor=3)
_model.fit(AD, DP, min_iter=30, n_init=50)
print(_model.ELBO_iters[-1])
-190779.74335041404
Check the model fitting¶
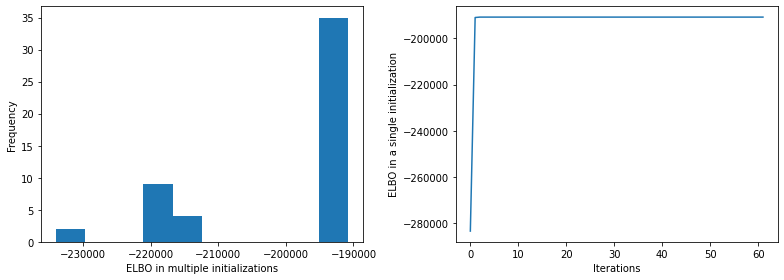
[6]:
fig = plt.figure(figsize=(11, 4))
plt.subplot(1, 2, 1)
plt.hist(_model.ELBO_inits)
plt.ylabel("Frequency")
plt.xlabel("ELBO in multiple initializations")
plt.subplot(1, 2, 2)
plt.plot(_model.ELBO_iters)
plt.xlabel("Iterations")
plt.ylabel("ELBO in a single initialization")
plt.tight_layout()
plt.show()
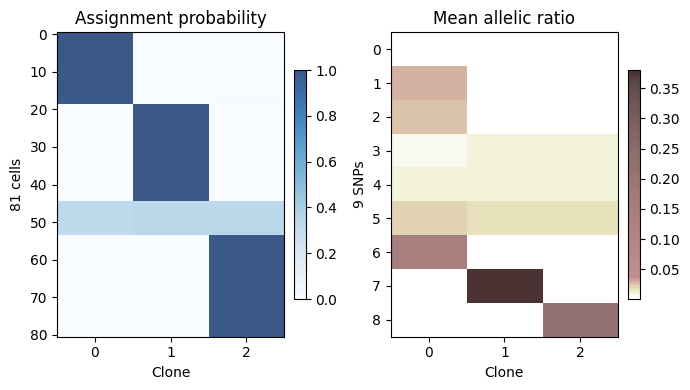
Visualize assignment probability and allele frequency¶
[7]:
# In mitochondrial, allele frequency is highly informative between 0.01 to 0.1,
# so we rescale the colour to give more spectrum for this region.
# You can design/choose your own colors from here:
# https://matplotlib.org/stable/tutorials/colors/colormaps.html
from matplotlib import cm
from matplotlib.colors import ListedColormap, LinearSegmentedColormap
raw_col = cm.get_cmap('pink_r', 200)
new_col = np.vstack((raw_col(np.linspace(0, 0.7, 10)),
raw_col(np.linspace(0.7, 1, 90))))
segpink = ListedColormap(new_col, name='segpink')
[8]:
from vireoSNP.plot import heat_matrix
fig = plt.figure(figsize=(7, 4), dpi=100)
plt.subplot(1, 2, 1)
im = heat_matrix(_model.ID_prob, cmap="Blues", alpha=0.8,
display_value=False, row_sort=True)
plt.colorbar(im, fraction=0.046, pad=0.04)
plt.title("Assignment probability")
plt.xlabel("Clone")
plt.ylabel("%d cells" %(_model.n_cell))
plt.xticks(range(_model.n_donor))
plt.subplot(1, 2, 2)
im = heat_matrix(_model.beta_mu, cmap=segpink, alpha=0.8,
display_value=False, row_sort=True)
plt.colorbar(im, fraction=0.046, pad=0.04)
plt.title("Mean allelic ratio")
plt.xlabel("Clone")
plt.ylabel("%d SNPs" %(_model.n_var))
plt.xticks(range(_model.n_donor))
plt.tight_layout()
plt.show()
# plt.savefig("you_favorate_path with png or pdf")
Diagnosis¶
We are using multiple initializations via n_init and choose the one with highest ELBO. However, this doesn’t guarantee to be the global optima. To double check it, we can run the same scripts multiple time (without fixed random seed), and check if the same (best) ELBO is found.
If yes, it is likely to be the global optima, otherwise, we need increase n_init, e.g., 300 for more search.
[9]:
n_init = 50
for i in range(3):
_model = BinomMixtureVB(n_var=AD.shape[0], n_cell=AD.shape[1], n_donor=3)
_model.fit(AD, DP, min_iter=30, n_init=n_init)
print("rerun %d:" %i, _model.ELBO_iters[-1])
rerun 0: -190779.74335041404
rerun 1: -190779.74335041404
rerun 2: -190779.74335041404
[ ]:
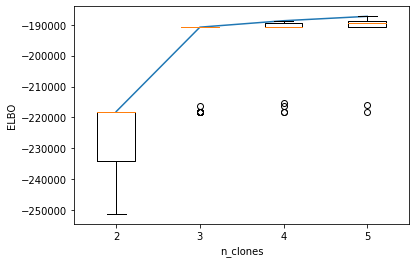
It is generally difficult to identify the number of clones, which is a balance between subclone resolution and analysis reliability. More clones maybe preferred, but there could be higher risk that the subclones are not genuine but rather technical noise.
Here, we could use ELBO for different number of clones as an indictor for model selection. However, this is still imperfect. One empirical suggestion is to choose the n_clones when ELBO stops increasing dramatically, for example in the case below, we will pick 3 clones.
[10]:
n_init = 50
n_clone_list = np.arange(2, 6)
_ELBO_mat = []
for k in n_clone_list:
_model = BinomMixtureVB(n_var=AD.shape[0], n_cell=AD.shape[1], n_donor=k)
_model.fit(AD, DP, min_iter=30, n_init=n_init)
_ELBO_mat.append(_model.ELBO_inits)
[11]:
plt.plot(np.arange(1, len(n_clone_list)+1), np.max(_ELBO_mat, axis=1))
plt.boxplot(_ELBO_mat)
plt.xticks(np.arange(1, len(n_clone_list)+1), n_clone_list)
plt.ylabel("ELBO")
plt.xlabel("n_clones")
plt.show()
[ ]:
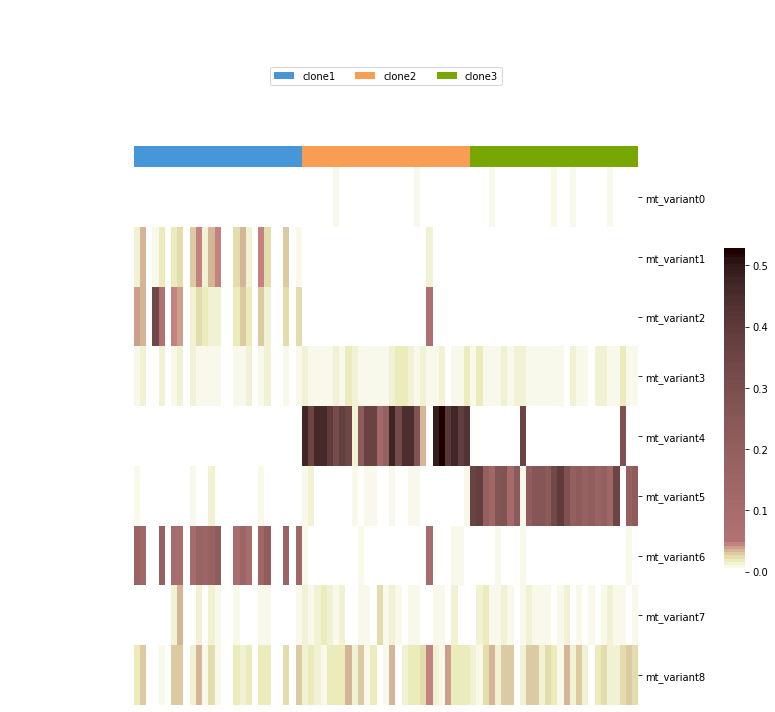
Visualization¶
If you want to visualise the raw allele frequency with annotation of cells, you may consider seaborn.clustermap. We also wrap this function here as vireoSNP.plot.anno_heat for quick use.
[12]:
mtSNP_ids = ['mt_variant%d' %x for x in range(AD.shape[0])]
cell_label = np.array(['clone1'] * 27 + ['clone2'] * 27 + ['clone3'] * 27)
id_uniq = ['clone1', 'clone2', 'clone3']
[13]:
vireoSNP.plot.anno_heat(AD/DP, col_anno=cell_label, col_order_ids=id_uniq,
cmap=segpink, yticklabels=mtSNP_ids)
[13]:
<seaborn.matrix.ClusterGrid at 0x7fd475067eb0>
[ ]: